This is the second in a series of posts I began this summer and didn’t have time to finish.
Edit 2011/01/17: I finally noticed and fixed a small glitch in the raw command line I was using, probably caused by my use of “<>” in the example text. Sorry, folks.
We all know how it feels, right? You’ve been coding away happily, decide it’s time to push everything to your version control system of choice, type out a brilliant commit message, and hit Enter–only to find out five seconds later that you just committed something you shouldn’t have committed and everyone else is in trouble when they happen to update. What do you do now?
Well, if you have a decent version control system (VCS) such as Mercurial or Git, you take advantage of the built-in undo or undo-like command and simply undo your commit. Most modern distributed VCS’s have one, giving you at least one chance to fix your work before you hand it out for everyone else to use. However, some less-than-fun VCS’s make it difficult (mostly because they’re designed to keep the information you give them forever, warts and all, which is not a bad thing). However, it’s still usually possible to fake an undo.
Take Subversion, for example. It’s not my favorite VCS for reasons I may go into later, but it’s still a very solid one if you like and/or need the centralized type of thing. I used it a lot this summer while doing an internship with Sentry Data Systems, and I found myself needing to rollback a commit on some files a time or two. So, I did some research.
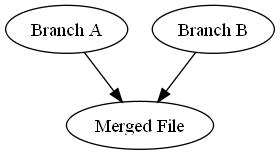
Arguably the best way to rollback a commit in Subversion is to perform what they call a reverse-merge. In a normal merge, you take two versions of a file, compare them to figure out what’s different between them, and then create a new file containing the consensus content of both files (see picture to the right).
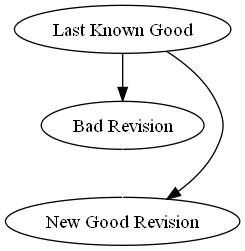
In a reverse-merge, you figure out what changes were made in a given revision, and then commit a new revision that reverts all of those changes back to the version before you committed your bad revision (see picture to the right), which means your bad revision will be simply skipped over as far as your comrades are concerned. Subversion provides a simple series of commands for this:
svn merge -c -{bad revision number} [{files to revert}]
Here’s how it works.
- The
svn mergeportion of the command basically tells Subversion you want to merge something. svn is the command-line tool to interact with a Subversion repository, and merge…well, you get the idea. -c -{bad revision number}tells Subversion that we want to work with the changes related to the revision numbered{bad revision number}. In this case, since we’re passing in a negative sign in front, we’re saying we want to remove those changes from the working directory. If you left out that negative sign, you’d actually pull the changes from that revision into the current working directory, which is usually only useful if you’re cherry-picking across branches. Whether that’s a good idea or not is left to the reader.[{files to revert}]are an optional list of files to undo changes in. Basically, if you pass in a list of files here, only those files will have their changes from revision–any other files changed in that revision will not be affected.
That’s pretty much it. Once you run that command (assuming there are no conflicts in your merge), you will be able to simply commit (with a helpful commit message, of course) and everything will be back to normal–your comrades in arms will be able to keep working without the overhead of your bad commit cluttering up their working environment, which is always a good thing.